13. HIRIPI L., DEVINOY E., RAT P., BARANYI Mária FONTAINE,M.L., BÕSZE Zsuzsanna: Characterization of a rabbit kappa-casein gene polymorphism. (A nyúl kappa-kazeingén polimorfizmusának vizsgálata)

13. Hiripi
érk: 97.11.21.

MBK, Állatgenetikai Int., 2100 Gödöllő, Szent-Györgyi A.u. 1.
Labor. Biologie Cellulaire et Moléculaire, Inst.Natl.Recherche Agronomique, 78 352 Jouy en Josas Cedex, France
INTRODUCTION
Caseins are a family of proteins which is composed of four to five members depending on species. Their encoding genes are closely located in the genome. The k-Cas gene has been shown to be unrelated to the other casein genes but to be related to a fibrinogen gene. Like fibrinogen, k-Cas can be cleaved by trypsin. The cleavage of k-Cas by enzymes or heat induces milk clotting. k-Cas is thus important to maintain the micellar structure of milk. Genetic studies have shown that higher k-Cas content were associated with smaller micellar size. In the bovine such intraspecies variations have been observed due to the k-Cas gene polymorphism. In this species, at least 6 alleles have been already described. Similar results were obtained for the ovine and caprine.
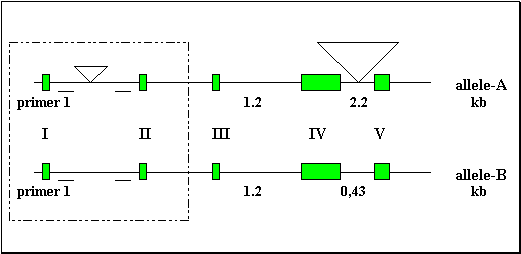
In the rabbit, four milk protein genes have been described and some RFLP of these genes described. The rabbit kappa-casein gene has been previously cloned and studied (fig1.) For the rabbit k-Cas gene, RFLP have been observed in DNA from white New Zealand rabbit digested with Hind III. Fragments of 4.7 kb, or 2.4 kb were observed. Rabbits showing a single 4.7 kb band, similar in size with the fragment obtained from the cloned gene, were thus homozygous and we designed the allele they harbour as the A allele. Other rabbits were homozygous for another allele that we designed as the B allele (B/B rabbits). Finally some rabbits were heterozygous for the A and B alleles (A/B rabbits).
Results described below show that the B allele is less frequent than the A allele, that the B allele is missing one half LINE sequence in the fourth intron and that part of the poly(T) track of an inverted LINE sequence is also lacking in the first intron. This allele is however expressed in the mammary gland of lactating rabbits to the same levels as the A allele.
Gene frequencies of two rabbit k-Cas alleles
Two independant populations of New Zealand rabbits were studied. The B allele is less frequent (28.4 %) than the A allele (71.6 %) (p<0.002, X2 test).
Localization of the polymorphism in the rabbit k-Cas gene
The RFLP described above has been observed with the Hind III enzyme, which cuts the cloned gene in the third intron and in its 3' flanking region. In the B allele, the sequences corresponding to the two Hind III sites could be altered by punctual mutations. Alternatively, the sequence corresponding to these sites could be conserved between both alleles and the length of the Hind III fragment defined by these two sites could be modified due to insertion/deletion events, in the third intron, in the fourth intron, or in the 3' flanking region of the gene. To study this polymorphism further, the lengths of the introns of k-Cas gene in two B/B rabbits, were evaluated after amplifications of DNA, using primers located in the exons and bordering the introns. The length of the fourth intron was only 0.43 kb for B/B rabbits instead of 2.2 kb as obtained for the B/B rabbits (fig2.). This 1.77 kb difference in the length of the fourth intron could partly explain the 2.4 kb difference observed between the 2.3 and 4.7 kb Hind III fragments obtained respectively from B/B and A/A rabbit genomic DNA The remaining difference between the Hind III fragments of the A and B allele is probably due to an alteration of the Hind III site which has been located at 1.1 kb in the 3' flanking region of the A allele.
The length of the first intron was shorter in the B/B rabbits studied (1.85 kb) than in the A/A rabbits (1.95 kb) (fig3.). The B allele thus harbours first and fourth introns which are respectively 100 bp and 1.77 kb shorter than in the A allele.
Sequences of the regions which appear different between the two genotypes
We compared the sequences of the 2 alleles, in regions which appear different in the first and the fourth introns. DNA amplified from the genomic DNA of an A/A rabbit. Sequences of the A and B alleles have been aligned. Downstream from the first exon, at position 315 a stretch of 106 nt present in the A allele is absent in the B allele. This stretch of 106 nt includes a serie of 25 (T) close to its 3' end. These 25 (T) are flanked 19 nt further downstream by a (GA)8C(AG)3 sequence. That microsatellite sequence present in the A allele is at the border of the rearranged region between the A and B alleles and is replaced by a (GA)9 sequence in the B allele. Over a 377 nt region located downstream from this rearranged region of the first intron, both A and B allele sequences exhibit 70.2% similarity with an inversed rabbit LINE sequence.
Downstream from the fourth exon, at position 350, a large sequence present in the A allele is absent in the B allele. Only the 3' end of this sequence has been deter-mined. It showed 79 % similarity with a rabbit LINE sequence over a 235 nt region and its 3' is characterized by the sequence (CT)3G(TC)7 followed 19 nt downstream by a stretch of 25(A). The 3'end of the sequence present in the fourth intron of the A allele, including the 19 nt, is exactly inverse and complementary to the sequence found in the rearranged region of the first intron.
Sequences of the mRNA encoded by the two alleles
The structure of the introns of the two alleles of the rabbit k-Cas gene being largely different, we decided to study also the sequences of the mRNA encoded by each allele. RNA extracted from the mammary glands of lactating rabbits homozygous for either allele were reverse transcribed and the resulting cDNA amplified. The amplification products were directly sequenced. Only two silent nucleotide substitutions were found between the two alleles in the coding region and three modification in the 3' untranslated region.
Comparison of k-Cas mRNA accumulations and milk concentration in the mammary glands of lactating rabbits having different genotypes
No signifiant variations of k-Cas mRNA concentrations in total RNA were observed between A/A, B/B or A/B rabbits. Milk was collected from A/A, A/B or B/B rabbits. Their protein concentrations were not significantly different. The genetic variation occurring at the k-Cas gene thus didn't largely alter either milk composition.
CONCLUSIONS
The present study describes an allele of the rabbit k-Cas gene, which has been identified in two independent populations of rabbits. This allele is less frequent than the allele previously described. It couldn't be associated with any special phenotype. The coding region of this allele differs from that of the more frequent allele by silent mutations so that both alleles encode identical proteins. Furthermore the accumulation levels of mRNA transcribed from both alleles, in the mammary gland of lactating rabbits homozygous for each allele, are similar. The influence of the k-Cas genotype over agronom-ical or reproductive performances of the rabbit is now under study in different populations of rabbits. k-Cas alleles could serve as a marker and allow us to follow the genetic diversity of those rabbit populations.
REFERENCES
Baranyi, M.; Aszodi, A.; Devinoy, E.; Fontaine, M. L.; Houdebine, L. M., and Bosze, Z. Structure of the rabbit kappa-casein encoding gene: Expression of the cloned gene in the mammary gland of transgenic mice. Gene. 1996 26; 174(1):27-34.
Baranyi, M.; Brignon, G.; Anglade, P., and Ribadeaudumas, B. New data on the proteins of rabbit (Oryctolagus cuniculus) milk. Comparative Biochemistry and Physiology B - Biochemistry & Molecular Biology. 1995
Hardison, R. C. and Printz, R. Variability within the rabbit C repeats and sequences shared with other SINES. Nucleic Acids Res. 1985; 13:1073-1088.
Leveziel, H.; Rodellar, C.; Leroux, C.; Pepin, L.; Grohs, C.; Vaiman, D.; Mahe, M. F.; Martin, P., and Grosclaude, F. A microsatellite within the bovine kappa-casein gene reveals a polymorphism correlating strongly with polymorphisms previously described at the protein as well as the DNA level. Animal Genetics. 1994 25(4):223-228.
Perez, M. J.; Leroux, C.; Bonastre, A. S., and Martin, P. Occurrence of a LINE sequence in the 3' UTR of the goat alpha(s1)-casein E-encoding allele associated with reduced protein synthesis level. Gene. 1994 147(2):179-187.
Fig 1.
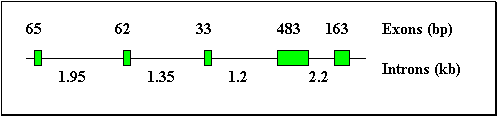
Fig 2.
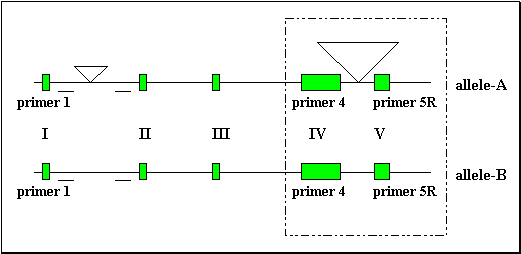
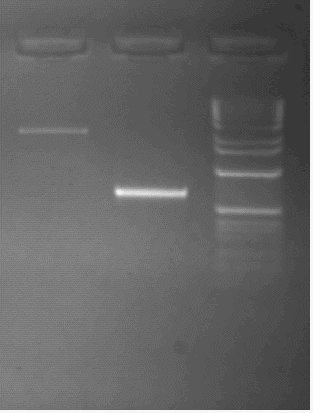
Fig 3.